Nanopore Sensing
Nanopore sensors enable rapid, portable, and relatively inexpensive DNA sequencing. But they can do so much more! We are exploring new and non-traditional ways to use commercial nanopore sensor arrays for bespoke molecular sensing, synthetic biology and proteomics.
Molecular tagging with Porcupine
London Calling 2019: Katie Doroschak lightning talk from Oxford Nanopore on Vimeo.
What are molecular tags? Molecular tags label physical objects using DNA or other molecules instead of conventional labels like RFID tags, QR codes, barcodes, and stickers. Molecular tags can be useful in cases where the tag needs to be small, lightweight, or invisible, or for tagging things that are too flexible or numerous.
Porcupine’s approach to molecular tagging is portable, quick to write and read new tags, and usable outside a lab setting, making it more accessible than existing molecular tagging methods. To write new tags, we combine tiny amounts of pre-prepared DNA solutions (molecular bits, or molbits), and to read tags, we use nanopore sequencing devices.
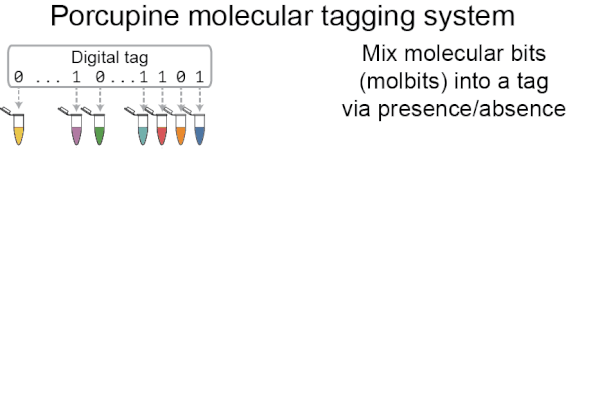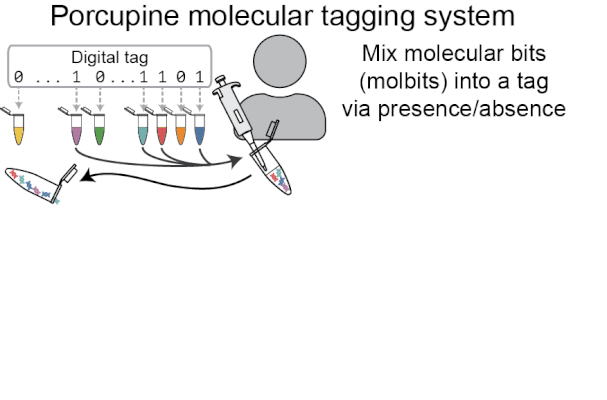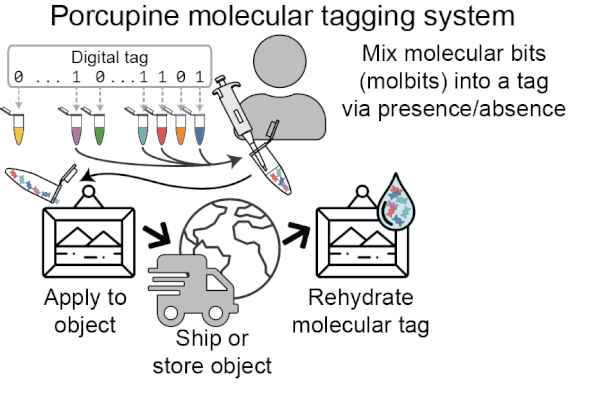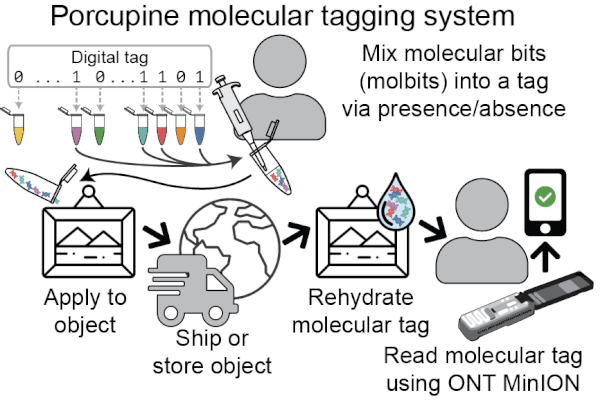
Porcupine starts with a digital tag, which is converted into a molecular tag using presence and absence (i.e., if a bit is set to 1, it is added to the molecular tag; if it is 0, it is left out). The tag is then prepared for nanopore sequencing and dehydrated in order to extend the tag’s shelf life, decrease readout time, and make tags robust to environmental contamination. When it’s ready to be read again, the tag is rehydrated and loaded onto a MinION flowcell. After sequencing for a brief period of time, we classify molbits directly from the raw nanopore signal, avoiding basecalling. We then identify the bits that are present and recover the digital tag.
More details on how we design the molecular tags, encode/decode data, and classify the molbits can be found in the publication.
Additional resources:
- Preprint
- Presentation at DEF CON Biohacking Village 2020 - YouTube
- GitHub
- Featured on Bioinformatics Chat podcast
Sensing protein barcodes (NanoporeTERs)
London Calling 2019: Jeff Nivala lightning talk from Oxford Nanopore on Vimeo.
NanoporeTERs: Nanopore-addressable protein Tags Engineered as Reporters Genetically encoded reporter proteins are a cornerstone of molecular biology. While they are widely used to measure many biological activities, the current number of uniquely addressable reporters that can be used together for one-pot multiplexed tracking is small due to overlapping detection channels such as fluorescence. To address this, we built an expanded library of orthogonally-barcoded Nanopore-addressable protein Tags Engineered as Reporters (NanoporeTERs), which can be read and demuxed by nanopore sensors at the single-molecule level. By adapting a commercially available nanopore sensor array platform typically used for real-time DNA and RNA sequencing (Oxford Nanopore Technologies’ MinION), we show direct detection of NanoporeTER expression levels from unprocessed bacterial culture with no specialized sample preparation. These results lay the foundations for a new class of reporter proteins to enable multiplexed, real-time tracking of gene expression with nascent nanopore sensor technology.
News
-
September 24, 2021
Check out our review on protein sequence analysis using nanopore sensors in Cell iScience
-
August 20, 2021
NanoporeTERs are covered by GenomeWeb! Prof. Jeff Nivala is quoted. New Protein Reporters for MinIon Sequencer Point Toward Route for Single-Molecule Proteomics
-
August 12, 2021
NanoporeTERs paper is published in Nature Biotechnology! NanoporeTERs enable more direct monitoring of intra-cellular activity with a mix of synthetic biology, nanopore sensing, and machine learning. So many possibilities in sensing, better data for ML, etc. Check out a nice UW Allen School news article about the work :)
-
November 03, 2020
Our molecular tagging system, Porcupine, is now published in Nature Comms! Our DNA-based tags can be assembled with minimal lab resources (4+ billion possible tags!), applied onto the surface of physical objects that you want to track, and are decodable with a portable nanopore sensor in just a few seconds.
-
April 08, 2020
Katie was interviewed for a podcast about Porcupine – Bioinformatics Chat by Roman Cheplyaka. The episode should be out by the end of the month!
People
Jeff Nivala
Katie Doroschak

Nick Cardozo
Karen Zhang

Aerilynn Nguyen
Karin Strauss
Affiliate Professor








